Molecular Structure Collection (page 22)
"Molecular Structure: Unlocking the Secrets of Life's Building Blocks" From anaesthetics inhibiting ion channels to antidepressant molecules
All Professionally Made to Order for Quick Shipping
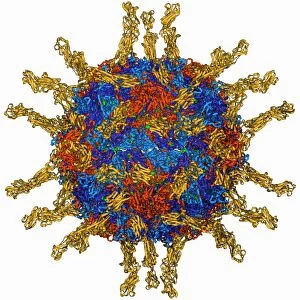
"Molecular Structure: Unlocking the Secrets of Life's Building Blocks" From anaesthetics inhibiting ion channels to antidepressant molecules, the intricate world holds endless wonders. The C015 / 6718 anaesthetic molecule delicately interacts with ion channels, altering their function and providing relief from pain. Meanwhile, Amitriptyline, an antidepressant molecule, works its magic by modulating neurotransmitters in our brains. In the realm of immunity, Immunoglobulin G antibody F007 / 9894 stands tall as a defender against pathogens. Its unique structure allows it to recognize and neutralize foreign invaders effectively. On another front, DNA artwork showcases the elegance and complexity that underlies all life forms on Earth. Creatine amino acid molecule fuels our muscles during intense physical activities while nanotube technology revolutionizes various industries with its exceptional properties. These tiny tubes hold immense potential for advancements in medicine and materials science alike. Zinc fingers bound to a DNA strand demonstrate how proteins can precisely interact with genetic material. This interaction plays a crucial role in gene regulation and expression. Carbon nanotubes take center stage once again as they exhibit remarkable strength and conductivity at the nano-scale level. Oxytocin neurotransmitter molecule reminds us of love's powerful influence on human connections—its presence promotes bonding between individuals. Manganese superoxide dismutase enzyme F006 / 9423 safeguards our cells by combating harmful free radicals that contribute to aging and disease. Even viruses have their own molecular structures; SARS coronavirus protein represents one such example—a key player in viral replication within host cells. Conceptual artwork further explores nanotube technology's limitless possibilities—the fusion of imagination and scientific innovation knows no bounds here. As we delve deeper into understanding molecular structures, we unravel nature's blueprint for life itself—one atom at a time. These captivating glimpses into the microscopic world remind us of both the fragility and resilience found within the building blocks of existence.