E Coli Collection
"E. Coli: Unveiling the Microscopic World of a Notorious Bacterium" In the realm of microscopic organisms, one name stands out - E. Coli bacterium
All Professionally Made to Order for Quick Shipping
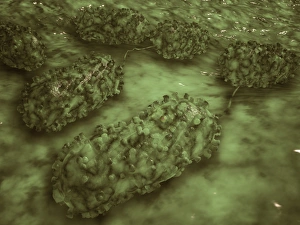
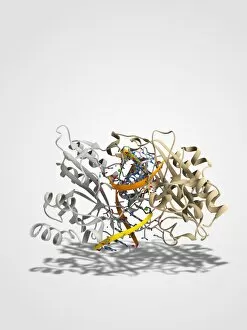
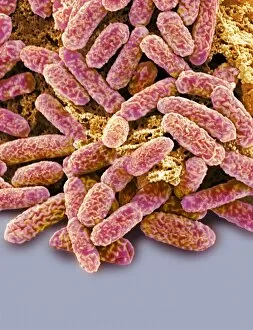
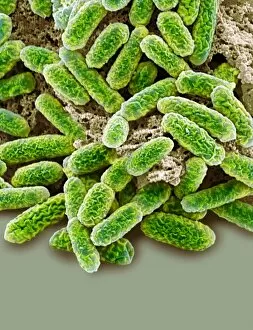
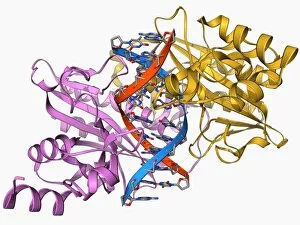
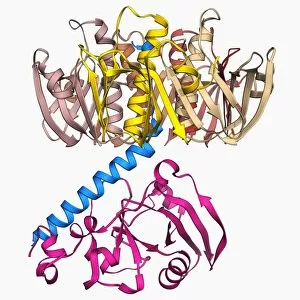
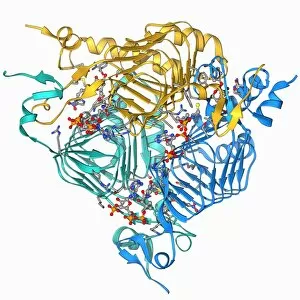
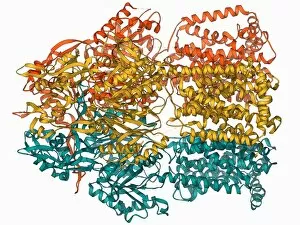
"E. Coli: Unveiling the Microscopic World of a Notorious Bacterium" In the realm of microscopic organisms, one name stands out - E. Coli bacterium. With its distinctive rod-shaped appearance, this tiny creature has captured the attention of scientists and researchers worldwide. Through advanced imaging techniques such as TEM (Transmission Electron Microscopy) and SEM (Scanning Electron Microscopy), we can now explore the intricate details of E. Coli bacteria like never before. One fascinating discovery is that these bacteria are not confined to their natural habitat but can also be found in unexpected places, even on our beloved mobile phones. SEM images reveal clusters of E. Coli bacteria thriving on these devices, reminding us of the omnipresence and adaptability of these microorganisms. A closer look at a group of Escherichia coli bacterial cells reveals their remarkable structure and organization. Known simply as E. coli, they form colonies with individual cells tightly packed together, creating an impressive sight under the microscope. Delving deeper into their molecular world, we encounter EcoRV restriction enzyme molecules and Endonuclease IV molecules – key players in DNA manipulation processes within E. coli cells. These enzymes play crucial roles in genetic engineering experiments by cutting or modifying DNA sequences. The EcoRV restriction enzyme molecule C014/2117 takes center stage among its counterparts due to its unique properties and potential applications in biotechnology research. Scientists harness this molecule's ability to cleave specific DNA sequences precisely for various purposes like gene cloning or studying genetic mutations. As we continue our exploration through microscopic lenses, it becomes evident that understanding E. coli goes beyond mere curiosity; it holds immense significance for both scientific advancements and public health concerns alike. By unraveling its secrets at a cellular level, we pave the way towards combating infections caused by this notorious bacterium more effectively.